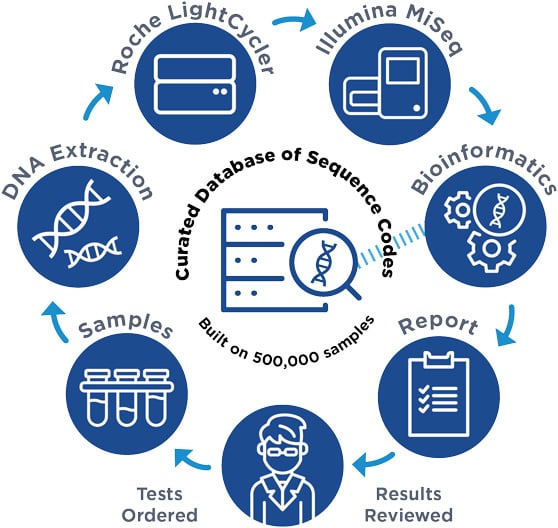
The MicroGenDX NGS Process – Perfected by Experience and Continuous Quality Improvement
Quality nucleic acid extraction
Nucleic acid extraction from multiple specimen types using tailored proprietary techniques that include concentrating cellular material, mechanical lysis, and removing organic inhibitors and human host DNA. Every specimen is processed with a positive and negative extraction control to assure quality.
Illumina MiSeq sequencing process
Illumina is the leading DNA sequencing platform, offering the lowest error rate of < 0.4%. Illumina’s 16S and ITS target gene deep sequencing afford high sensitivity, high discovery power, and high mutation resolution, with three positive and one negative control run alongside every specimen throughout the entire sequencing process to ensure the highest sensitivity and specificity possible.
Superior, curated, bacterial and fungal sequence database
Over 500,000 samples processed using a validated microbial database of over 50,000 referenced microbes, ensuring representation of every possible infective organism, resulting in 12 years of molecular testing quality and reliability.
2009-2020 CAP proficiency results: NGS identification to the species level
| Species panel | Test frequency | Number of blinded samples per test | Total number of blinded specimens | Proficiency score |
|---|---|---|---|---|
| IDR | 3 times per year for 7 years | 5 | 105 | 98.1% |
| MRS-5M | 3 times per year for 12 years | 5 | 180 | 100% |
| F1 | 3 times per year for 12 years | 5 | 180 | 98.30% |
| D8 | 3 times per year for 12 years | 5 | 180 | 99.40% |
| VRE | 2 times per year for 12 years | 2 | 50 | 100% |
| GIP | 3 times per year for 4 years | 5 | 60 | 100% |
| HC-6 | 3 times per year for 3 years | 5 | 45 | 100% |
| TVAG | 2-3 times per year for 2 years | 3 | 21 | 90.50% |
| Alternate | 3 times per year for 12 years | 6 | 216 | 100% |
| See species panel key | Blinded specimen total: 1,037 | Mean accuracy rate: 99.2% | ||