Case studies for qPCR+NGS
Chronic sinusitis requiring dual antibiotic therapy
A 57-year-old male with longstanding chronic sinusitis and history of prior endoscopic sinus surgery presented with an exacerbation of his chronic sinusitis. A sinus aspirate was obtained and MicroGenDX next-generation DNA sequencing performed. This revealed medium quantities of bacterial DNA. Three facultative anaerobic organisms were identified: Enterobacter cloacae, Corynebacterium segmentosum, and Staphylococcus epidermidis. The Enterobacter species was found to represent 61% of the bacterial DNA, and the Corynebacterium represented 34%, while S. epi represented only 2% of the DNA – confirming it to be a contaminant. To adequately treat the infection, and given the lack of oral antibiotic coverage with a single agent for both species, dual antibiotic therapy with both doxycycline and ciprofloxacin was used. The patient had complete resolution of infection with one 14-day course of therapy.
This case highlights two important advantages of next-generation DNA sequencing for microbial identification in patients with chronic sinusitis. First, although three bacterial species were identified, the quantitative nature of the results allowed for determination of the relative contribution of each species to the infection, including determining that S. epi was a contaminant. Second, by identifying more than one organism, the need for combined antimicrobial therapy was established, and a treatment with both ciprofloxacin and doxycycline resulted in rapid resolution of infection. The selection of ciprofloxacin and doxycycline would certainly not have occurred as empiric therapy.
Patient with urinary tract infection
An 82-year-old woman with a history of recurrent UTI, and who had been treated for E. coli, presented to the clinic with an acute episode.
A mid-stream urine sample was obtained and sent for standard urinalysis, C&S, PCR by Volente Laboratories and MD Labs, and NGS by MicroGenDX.
There was a high concordance between C&S and PCR-based techniques to detect a high load of E. coli (>105 per ml). All testing identified E. coli, but NGS was the only lab that detected the dominant pathogen as Bacteroides fragilis (50%). Based on the anaerobic B. frag and quinolone-resistant genes detected by NGS, the patient was switched to a targeted antimicrobial therapy using clindamycin for a total of 14 days. By the fifth day, the patient’s symptoms subsided, and upon follow-up on day 14, she had no symptoms. View study
Fungal infection in mastoid cavity
A 79-year-old male with a longstanding history of chronic ear disease has had multiple middle ear and mastoid surgeries, leaving him with a mastoid cavity and non-intact tympanic membrane. He developed an intermittent mastoid cavity infection typically treated with topical ciprofloxacin drops. He then presented with recurrent otorrhea. A sample of the mastoid cavity discharge was collected for MicroGenDX qPCR+NGS testing. This revealed the presence of Aspergillus tamarii — this result was available four days after collection of the specimen. No bacterial DNA was identified, confirming the fungal etiology of the infection. The ciprofloxacin drops were immediately discontinued and topical antifungals used instead with resolution of the infection.
This case demonstrates an important advantages of next-generation DNA sequencing over traditional culture technique, namely time-to-diagnosis. The slow growth of fungi in culture may require more than one week of incubation depending on the species.
Patient with prostate infection
A 44-year-old police instructor was placed on disability with chronic pelvic pain, and missed work for two years. A urine sample revealed E. coli, and after one month of antibiotics his symptoms seemed to resolve. Then he relapsed. His cultures were negative, and CT, scope, and MRI were normal. Consulting five urologists, none could help — he was referred to physical therapy, which made symptoms worse. MicroGenDX reported urine was negative, but semen showed Enterococcus resistant to all oral antibiotics. The patient was given Amikacin IM at 2, 4, 6, and 7 days, and his symptoms resolved.
Non-healing leg ulcer closed using MicroGenDX results
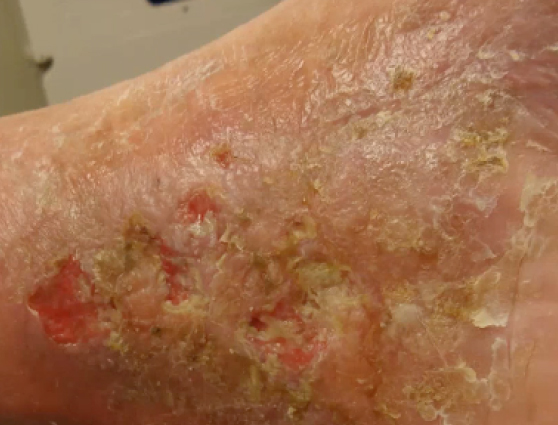
A 70-year old patient presented with a venous leg ulcer that had persisted for 15 years. The wound remained open despite treatment based on multiple culture results.
A MicroGenDX qPCR+NGS test revealed Pluralibacter gergoviae as the dominant species in the wound at 82%, with three other species detected as well:
S. epidermidis (6%)
E. coli (5%)/em>
A. vaginalis (2%)
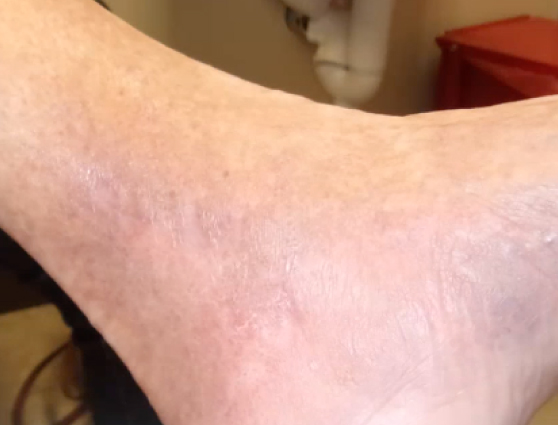
No fungal species were detected. The patient was then treated with a compound topical, and the wound closed within six weeks and has remain closed in episodic follow-ups.
Pluralibacter gergoviae is an opportunistic pathogen, and can be exceptionally resistant to antibiotics. Clinically, it has be recovered from respiratory samples, wounds, blood cultures, stools and urine. Pluralibacter gergoviae has also been isolated from environmental samples, coffee beans, refrigerated packed food, and various cosmetic products. The patient had been a restaurant owner for 17 years, handling refrigerated packaged food on a daily basis.
