The timely and reliable identification of causative microorganisms is critical to ensure the appropriate diagnosis and treatment of infectious diseases. Traditionally, microbial cultures have been utilized to identify causative pathogens and test their antimicrobial susceptibility.1 However, this approach may encounter serious challenges. Thus, antibiotic treatment suppresses pathogens and interferes with their culture, even if it does not completely eradicate them.2
Moreover, a number of microorganisms are difficult or impossible to culture. qPCR + next-generation sequencing (NGS) represents an innovative and reliable alternative for pathogen identification and diagnosis in cases where more traditional techniques fail, such as after antimicrobial treatment and for the diagnosis of fastidious microorganisms.3
Next-generation sequencing can reliably detect microorganisms even after the initiation of antimicrobial treatment. Generally, samples for microbial identification should be collected prior to the initiation of antimicrobial treatment. However, in certain instances, this specimen collection does not occur — for example, if empiric therapy is indicated or if the clinical situation is urgent and prevents immediate testing.
If pathogen identification becomes necessary after the initiation of antimicrobial treatment, the therapy will interfere with detecting microbial agents in culture because it will suppress microorganisms and limit their growth ability, leading to false-negative results. However, NGS does not require the microbial organisms to be fully viable for their successful identification.
Thus, NGS gives researchers the flexibility to identify pathogens, even if the collection of specimens prior to antimicrobial therapy initiation fails or if a second specimen needs to be obtained after the therapy initiation.
The primary focus of NGS use is on the diagnostic process. Generally, testing should follow the clinical evidence, and if the clinical symptoms resolve, no test of cure is recommended; a second test is generally needed only if symptoms persist. However, if NGS is used as a test of cure, at least a week should elapse after the completion of antibiotic treatment to eliminate the risk of false-positive results.
Next-Generation Sequencing a Valuable Diagnostic Tool — Clinical Use Cases
There are various clinical scenarios in which NGS can be a highly valuable microbiological diagnostic tool, such as in patients with sepsis who have already initiated antimicrobial treatment or in patients with urinary tract infections or sinus infections that remain symptomatic after empirical antibiotic therapy.
Microbiological diagnosis of patients with sepsis who have already initiated antimicrobial treatment
In patients with sepsis, both the initiation of intravenous antimicrobial treatment and specimen collection for pathogen testing should occur within an hour of admission. In such urgent situations, specimen collection for pathogen identification may have to be performed after the initiation of antimicrobial treatment. The recently administered antimicrobial drug will be highly concentrated in the blood, hindering pathogen growth in culture. Fortunately, NGS will allow reliable determination of the microorganism.
Urinary tract infection (UTI) and persistent symptoms after empiric treatment
A more specific treatment is needed in patients with a UTI who have completed an empirical antibiotic therapy but remain symptomatic. However, the antibiotic therapy suppresses the pathogens, even if it does not eradicate them, hindering their ability to grow in culture. This problem is further exacerbated by the fact that antibiotics reach high concentrations in urine.
Next-generation sequencing is a diagnostic procedure of choice in this situation because it enables the identification of causative pathogens even after the completion of an antimicrobial course. An important aspect in this context is the significance of antisepsis and the prevention of contamination in the process of sample collection to eliminate the risk of false-positive results.
Sinus infection after unsuccessful empiric treatment
If a patient presents with sinus infection symptoms and the medical history does not indicate a risk of significant complications, the diagnosis may be confirmed with an X-ray, and sample collection for pathogen testing may be foregone to prevent patient discomfort. In such cases, the patient starts an empiric treatment, commonly with doxycycline, a bacteriostatic antibiotic suppressing bacterial growth rather than killing the bacteria.
However, sinus infections are often polymicrobial and may be resistant to treatment. If the patient still experiences symptoms such as pain and discharge at the end of the antibiotic treatment, the physician may decide to perform sample collection to identify the causative agents and start a specific antibiotic therapy. At that moment, NGS is a suitable strategy for pathogen detection because it can reliably identify microorganisms whose ability to grow in culture has been suppressed by the antimicrobial treatment.
The Capability of NGS to Detect a Wide Range of Microorganisms
Another important advantage of next-generation sequencing is that it enables researchers to identify a broad range of aerobic and anaerobic bacteria and certain fungi.4 Moreover, NGS can detect irregular, atypical bacteria that are challenging to, or won’t grow on, culture such as mycoplasmas and ureaplasmas (intracellular bacteria). Further, NGS can be performed using specimens from a number of different tissues, including urine, blood, saliva, sputum, bronchoalveolar lavage, respiratory swabs, tissue drainage, cerebrospinal fluid, synovial fluid, and heart valves.
MicroGenDX’s Innovative NGS Technology for Microbiological Diagnostics
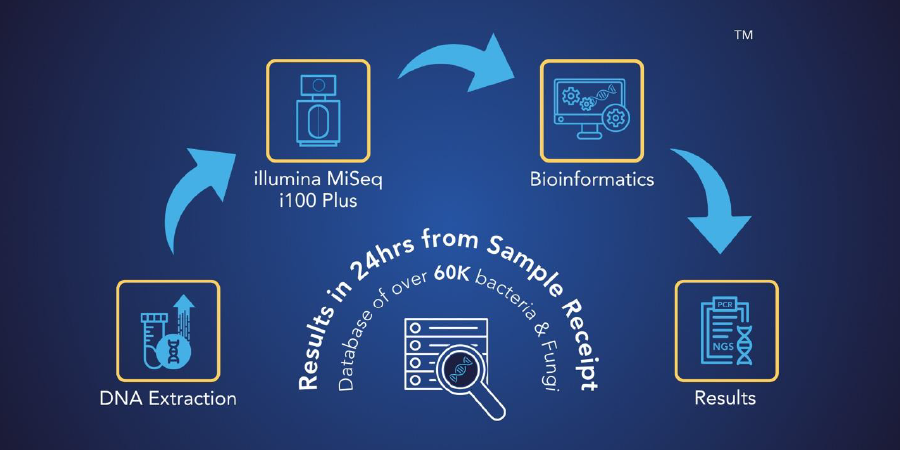
MicroGenDX is an innovative molecular diagnostic laboratory that offers reliable microbiological testing services utilizing our proprietary qPCR+ NGS technology. Our laboratory is CAP-accredited and CLIA-licensed and has been a trusted research partnership lab to the FDA, NASA, and the CDC.
We have over 13 years of 16S/ITS NGS testing quality and reliability and have processed over 500,000 qPCR+NGS samples. Moreover, we have developed a comprehensive proprietary bioinformatics system and curated database that ensure the precise detection of infectious diseases at high levels of sensitivity and specificity.
To learn more about MicroGenDX’s innovative microbial testing platform and to discuss your project with our expert team, contact us today at microgendx.com!
Resources
- Giuliano C, Patel CR, Kale-Pradhan PB. A Guide to Bacterial Culture Identification and Results Interpretation. P T. 2019 Apr;44(4):192-200. PMID: 30930604. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6428495/
- Hidden in plain sight: viable but non-culturable (VBNC) bacteria. https://microgendx.com/vbnc-viable-but-non-culturable-cells-dectection/
- Applications of Clinical Microbial Next-Generation Sequencing: Report on an American Academy of Microbiology Colloquium held in Washington, DC, in April 2015. Washington (DC): American Society for Microbiology; 2016. doi: 10.1128/AAMCol.Apr.2015. https://www.ncbi.nlm.nih.gov/books/NBK513764/
- Chen, P., Sun, W., & He, Y. (2020). Comparison of the next-generation sequencing (NGS) technology with culture methods in the diagnosis of bacterial and fungal infections. Journal of thoracic disease, 12(9), 4924–4929. https://doi.org/10.21037/jtd-20-930