MicroGenDX qPCR + NGS Facilitates Antimicrobial Stewardship for Challenging Infections

MicroGenDX Next-Generation DNA Sequencing (NGS) demonstrates higher sensitivity than culture — an ideal complementary tool for high-risk infection diagnosis that mitigates the antimicrobial resistance risks of empirical treatment. Strong clinical evidence supports the utility of NGS in diagnosing and treating complex and deep-seated infection, allowing healthcare providers to reduce the duration of treatment and avoid unnecessary, repeat courses of multiple antimicrobials.
Where other diagnostic technologies fail, MicroGenDX qPCR+NGS identifies relative abundance of pathogens to species level in just 24 hours from sample receipt
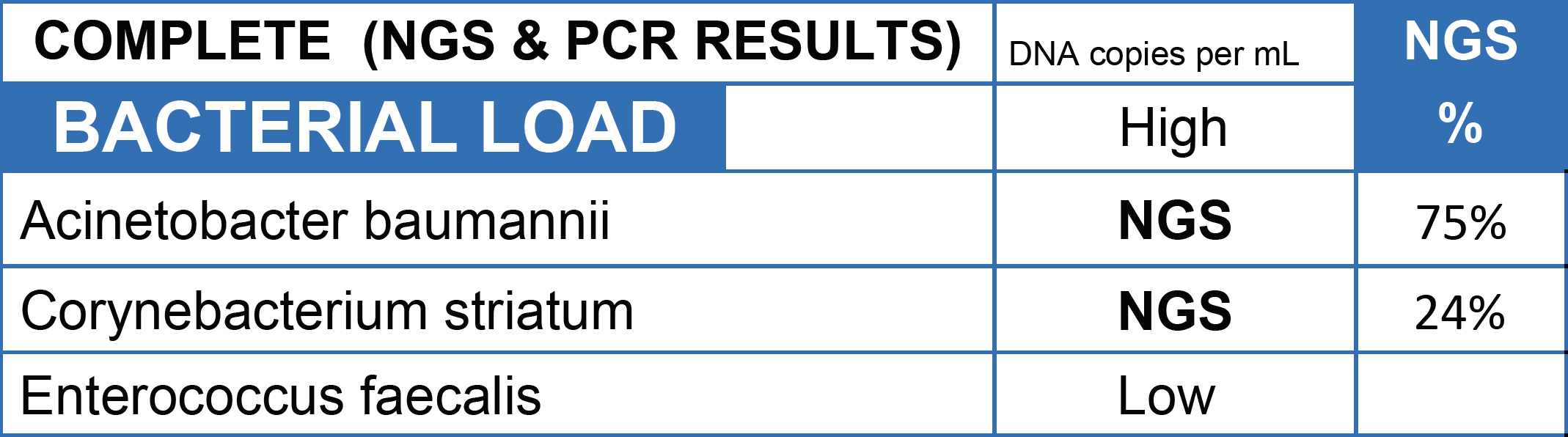
CASE STUDY: Culture returned mixed skin flora on a patient whose leg was on the verge of amputation due to a diabetic ulcer. MicroGenDX NGS detected Acinetobacter baumanii as the dominant pathogen, which can cause necrotizing infections and exhibits resistance to multiple antimicrobials. With treatment targeting A. baumanii, the patient's leg was saved with the appropriate antibiotic.
NGS identifies infective pathogens missed by culture in periprosthetic joint infections
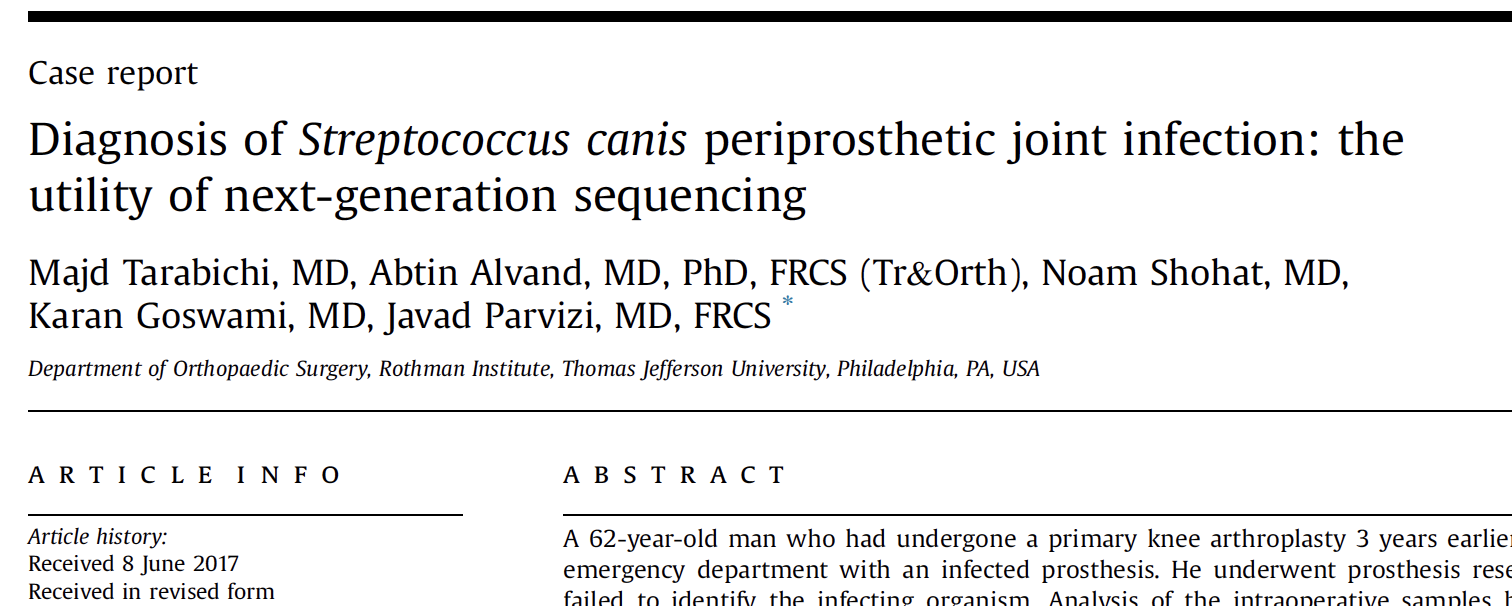
CASE STUDY: A 62 year old man presented with an infected prothesis from a primary knee arthroplasty three years prior. Next-generation sequencing identified the infective organism as Streptococcus canis, which cultures failed to detect. The patient was successfully treated and later underwent reimplantation arthroplasty. Read report
30% of E.coli UTIs are Bactrim-resistant
DOH STUDY: In a 2019 study by the NYC Department of Health, 30% of UTIs caused by E.coli were found to be resistant to Bactrim treatment, and some 20% were resistant to other common treatments. Read article
Detailed case discussions
The five case discussions below further illustrate the efficacy of MicroGenDX qPCR+NGS in combating AMR.
Case discussion #1: Rapid mycobacterial identification
| NGS Results | |
|---|---|
| Deep Tissue #1 | Mycobacterium smegmatis 34% Serratia marcescens 17% Streptococcus sanguinis 10% |
| Deep Tissue #2 | Serratia marcescens 28% Mycobacterium smegmatis 27% Staphylococcus epidermidis 8% |
| Superficial Tissue #1 | Serratia marcescens 84% Mycobacterium smegmatis 6% |
| Superficial Tissue #2 | Serratia marcescens 45% Mycobacterium smegmatis 38% Acinetobacter guillouiae 3% Staphylococcus epidermidis 2% Corynebacterium aurimucosum 2% |
| Hardware Swabs | Negative |
A 62-year-old female underwent spine revision surgery due to screw displacement. Culture of tissue obtained intraoperatively grew a nontuberculous mycobacteria (NTM) in five days, however a specific species was not identified. Next-generation sequencing (NGS) analysis reported the presence of Mycobacterium smegmatis within five days, providing confidence in the choice of targeted therapy. Culture then confirmed the NGS speciation, but only at day 35. Patient was placed on tigecycline and ciprofloxacin for six weeks, followed by suppressive antibiotics. A follow-up at 18 months confirmed no signs of recurrent infection.
Antibiotic treatment differs greatly between infections of different mycobacteria species, and pathogens must be distinguished from environmental contaminants. A broad anti-mycobacterial medication over many months is typically required to cure this type of infection.
To read more about Mycobacterium Smegmatis infection cases, view this article
Case discussion #2: Antibiotic de-escalation
| Culture Results | NGS Results | |
|---|---|---|
| Preoperative As-piration | Synovial fluid: Kocuria spp 67% | |
| Intraoperative Samples | Synovial fluid: Gram positive cocci Bone: No growth Tissue #1: Very light growth Staphylococcus spp Tissue #2: Very light growth Staphylococcus spp Tissue #3: Very light growth Gram positive cocci | Synovial fluid: Negative Tibia: Kocuria ocularis 93% Femur: Negative Tibia/Femur swabs: Negative |
Synovial fluid and intraoperative samples from a single stage revision knee arthroplasty grew in culture, and Staphylococcus spp was identified by MALDI-TOF. The patient was discharged on vancomycin and rifampin, and given a presumptive diagnosis of staphylococcal infection. NGS testing meanwhile revealed that only Kocuria ocularis was detected. Consultation with the Infectious Diseases team led to further investigation and further testing of cultured sample against a research-use-only MALDI-TOF database. It was confirmed that the sample contained Kocuria species. Antibiotic susceptibilities performed on the original culture allowed treatment change to penicillin in order to target Kocuria ocularis. At follow-up six months after surgery, the patient was doing well with improved knee function and no evidence of reinfection.
To read about other Kocuria infection cases, view this article
Case discussion #3: Correct microbial detection leads to effective initial treatment
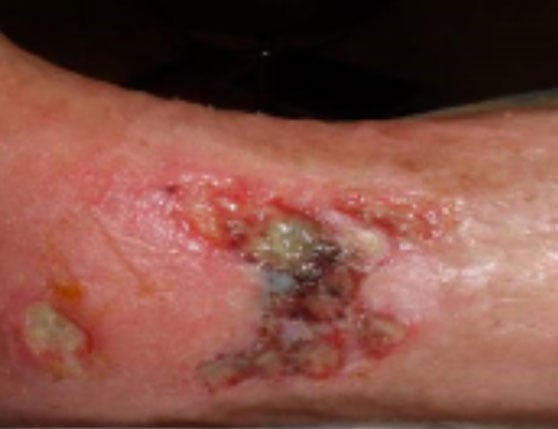
A frail older woman with a slowly improving leg wound which suddenly began to get worse. Her culture showed 1+ Pseudomonas aeruginosa and 1+ skin flora. Her MicroGenDX test showed that Pseudomonas was only 14% of the bacterial DNA load, accompanied by a colony of anaerobes — chiefly Bacteroides pyogenes at 43%, which is a significant clinical pathogen. Topical compounded antimicrobials targeting both pathogens effectuated marked wound recovery. Read more about this case
Case discussion #4: Ending an ineffective antibiotic acceleration cycle
A 68-year-old female with a one-year history of sinusitis presented to ENT with symptoms of sinusitis, having previously received multiple courses of antibiotic treatment based on culture.
MicroGenDX test report revealed six oral anaerobes, including Porphyromonas endodontalis and Prevotella oris, making up 74% of the bacterial load. CT showed unilateral maxillary sinus opacification and a ruptured periapical abscess of an underlying maxillary molar. Surgery was then performed following extraction of the tooth. Following surgery and additional treatment with clindamycin, her infection resolved.
This case highlights the superior ability of next-generation sequencing to detect the presence of anaerobes compared with culture. This allows for the identification of oral anaerobes suggestive of odontogenic sinusitis, which can then prompt the performance of a CT of the sinuses to detect the odontogenic source. Rapid identification of odontogenic sinusitis can then result in appropriate dental care to treat the source of the infection as well as help direct appropriate antimicrobial therapy. View NGS report
Case discussion #5: Rapid fungal identification saved a life
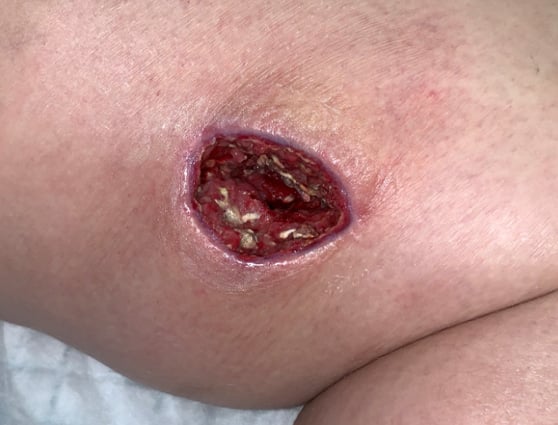
A young woman with type I diabetes, who had lost her kidney transplant a year previously, was hospitalized twice over six months with sepsis, the source of which remained a mystery. A large transplant surgery abdominal wound had been open for months, as had a heel pressure sore complicated by peripheral arterial disease, but both improved with better nutrition and regular wound care.
The patient then presented with sudden left leg pain, swelling, and the rapidly-ascending erythema characteristic of necrotizing infection. Fasciotomies of her lower leg were performed, and her left medial thigh was opened to drain what appeared to be an abscess. The muscle was perfectly pink and only clear, odorless fluid was found. This fluid then showed no growth on culture. Although wounds improved initially with NPWT, the patient didn’t look or feel better, and within a week the leg wound had grey/black tissue that appeared to be necrotic.
The same fluid was sent to MicroGenDX for DNA analysis (see full report), which detected Rhizopus — one of several fungal species that cause mucormycosis — a serious but rare fungal infection with a mortality rate of 50%. Early detection and treatment are critical for the best outcome, and the transplant team was notified immediately. Read study
Read more from Dr. Caroline Fife using MicroGenDX test results:
Pondering DNA Assay of Wounds: It only Matters When it Matters
At a Glance – When is a Dog NOT Man's Best Friend?
References
- Corona PA, Goswami K, Kobayashi N, Li W, et al. General Assembly, Diagnosis, Pathogen Isolation: Proceedings of International Consensus on Orthopedic Infections. The Journal of Arthroplasty. 34 (2019) S207eS214. https://doi.org/10.1016/j.arth.2018.09.0724.
- Tarabichi M, Shohat N, Goswami K, et al. Diagnosis of Periprosthetic Joint Infection: The Potential of Next-Generation Sequencing. J Bone Joint Surg Am. 2018 Jan 17;100(2):147-154.doi: 10.2106/JBJS.17.00434
- Asten SAV, La Fontaine J, Peters EJG, et al. The microbiome of diabetic foot osteomyelitis. Eur J Clin Microbiol Infect Dis. 2016 Feb;35(2):293-8. doi: 10.1007/s10096-015-2544-1
- Cheng J, Hu H, Fang W, et al. Detection of pathogens from resected heart valves of patients with infective endocarditis by next-generation sequencing. Int J Infect Dis. 2019;83:148-153. doi:10.1016/j.ijid.2019.03.007
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4232501/
- https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis
- https://www.cdc.gov/hicpac/recommendations/antibiotic-stewardship-statement.html
