The Limitations of Culture

“Many times these patients are culture negative. That is a big problem in our practice and in general orthopedics, is that we know they are infected, we see they are infected, but we can’t pin down the cause of the infection.”
— Dr. Jon E. Minter, DO, Arthritis & Joint Specialists, Alpharetta, GA
Why do the limitations of culture matter for periprosthetic joint infection (PJI)?
Sensitivity of routine culture in identifying infecting organisms in PJI is low, ranging from 39% to 70%. Negative culture PJI is associated with poorer outcomes, and the rate of treatment failure for culture negative PJI can be as high as 69%. Contrary to a widely held assumption, polymicrobial instead of monomicrobial infections make up the majority of PJI, but culture is not proficient at identifying multiple organisms in each sample. Incomplete treatment resulting in chronic infection may account for 70% of revision failures. The mortality and cost associated with PJI treatment are high, with a 5-year mortality rate of 25%, and average cost of $15,000 – $20,000.
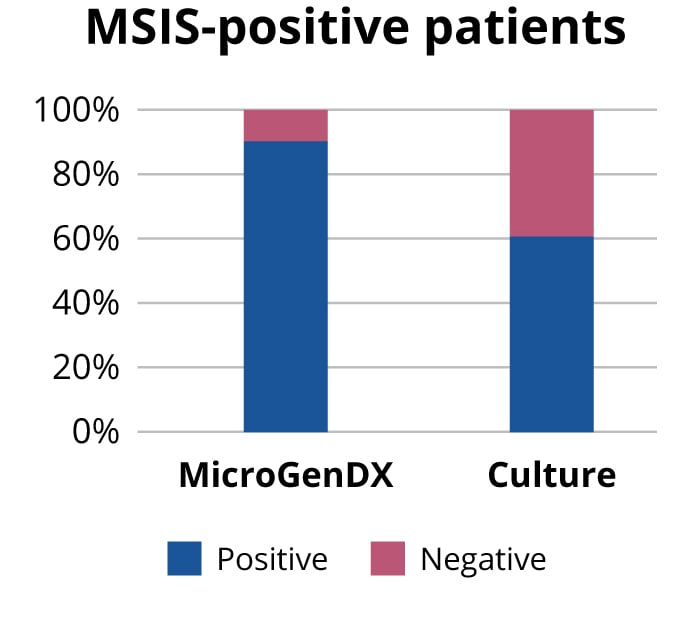
Instead of relying only on either culture or NGS, why not use both to maximize the probability of treatment success? NGS has high concordance with traditional culture, 96% based on a 2018 study published in the Bone and Joint Journal, and higher sensitivity than culture — some 90% vs 60% based on another Bone and Joint Journal published study (see references below). MicroGenDX is the only molecular testing lab that has published concordance data with PJI culture results.
Comparing C&S and qPCR + NGS
| Traditional Culture | MicroGenDX qPCR+NGS |
|---|---|
|
|
|
|
|
|
Appreciating the viable but noncultural (VNBC) bacterial state
Originally described by Rita Colwell’s group in 1982, it is now believed that any environmental stressor could send a bacterial community into this unculturable but still virulent state. Known human pathogens such as E. coli, E. faecalis, P. aeruginosa and S. aureus are all capable of entering and resuscitating from a VBNC state without losing their virulence. During the VBNC state, microbes maintain their cellular structure and biology, along with significant gene expression, but they are nonculturable by ‘standard’ laboratory methods. NGS can detect these VBNC bacteria.
C&S understates antimicrobial resistance
Culture has proven inadequate navigating in vivo and in vitro differentiated gene expression, enhanced pathogenicity in polymicrobial colonies, biofilm formation, and drug-resistant subpopulations. These shortcomings often result in unidentified antimicrobial resistance.
Resistance gene expression may be much higher in vivo than in vitro: a transcriptome analysis of P. aeruginosa in human infection and a variety of lab conditions found resistance genes had substantially higher expression in human infection versus culture. In addition, culture may underestimate antimicrobial susceptibility of a polymicrobial infection from two standpoints:
- Culture may flag polymicrobial infections as monomicrobial or dismiss them as contaminants
- Antibiotic sensitivity analysis is performed on single bacteria, not polymicrobial colonies
The relationship between S. aureus and P. aeruginosa is a well-documented instance of how polymicrobial infection can reduce antimicrobial susceptibility. S. aureus can be protected from tobramycin when growing with P. aeruginosa capable of producing Psl, and S. aureus components like SpA can enhance the antimicrobial resistance of P. aeruginosa biofilms.
Biofilms that culture cannot replicate can also dramatically reduce antimicrobial susceptibility due to these factors:
- The limitations of antibiotic diffusion through the biofilm matrix
- Transmission of resistance genes throughout the entire biofilm community
- Physiological changes in microbial cells — such as reduced metabolic and growth rates — due to the nutrient limitations of the biofilm environment
In addition, small heteroresistant subpopulations of cells can replicate in antibiotics despite the majority population being susceptible. Similarly, persister and VBNC cells are drug-tolerant phenotypes whose stress-induced dormant state allows them to survive the presence of antibiotics — after which they can rapidly resuscitate when surrounding conditions improve.
References
- Ehrlich G D et al. Culture Negative Orthopedic Biofilm Infections, Springer Series on Biofilms 7. 2012. doi.org/10.1007/978-3-642-29554-6_1
- Tan T L, Kheir M M, et al. Culture-Negative Perioprosthetic Joint Infection, An Update on What to Expect, JBJS Open Access:September 25, 2018 - Volume 3 – Issue 3 - p e0060 doi: 10.2106/JBJS.OA.17.00060
- Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance, NPJ Biofilms and Microbiomes, 2017
- Penterman J, Nguyen D, Anderson E, et al. Rapid evolution of culture-impaired bacteria during adaptation to biofilm growth. Cell Reports. 2014 Jan;6(2):293-300. DOI: 4.1016/j.celrep.2013.12.019
- Baron, E J, Miller J M, et al. A Guide to Utilization of the Microbiology Laboratory for Diagnosis of Infectious Diseases: 2013 Recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin Infect Dis. 2013 Aug;57(4):e22-e121. doi: 10.1093/cid/cit278.
- Tarabichi M, Shohat N, Goswami K, et al. Diagnosis of Periprosthetic Joint Infection: The Potential of Next-Generation Sequencing. J Bone Joint Surg Am. 2018;100(2):147-154.
- Tarabichi M, Shohat N, et al. Can next generation sequencing play a role in detecting pathogens in synovial fluid? The Bone & Joint Journal. 2018 Feb;100-B(2):127-133. doi: 10.1302/0301-620X.100B2.BJJ-2017-0531.R2
