A Case for NGS
NGS identifies Kocuria missed by MALDI-TOF in joint infection
A 77-year-old male with history of type 2 diabetes mellitus, hypertension, and hyperlipidemia presented to clinic with pain and stiffness to his right knee. Surgical history included a right TKA and subsequent revision surgery. Biomarkers from synovial fluid and serum were consistent with infection.
Synovial fluid and intraoperative samples were sent for culture MALDI-TOF and MicroGenDX NGS identification. Culture MALDI-TOF isolated very light growth Staphylococcus spp. and gram positive cocci. However, MicroGenDx NGS identified Kocuria ocularis to be the main culprit. A research MALDI-TOF database confirmed NGS identification of Kocuria ocularis. Patient treatment changed from vancomycin to penicillin, and patient was recovering well at 6-month follow-up.
The reference database used for MALDI-TOF did not contain Kocuria ocularis. An inability to identify an organism in this manner, along with the morphologic similarity to some coagulase-negative Staphylococcus (CoNS) species, led to the inaccurate reporting of the sample and initial implementation of incorrect antibiotic treatment.
NGS quickly identifies Mycobacterium smegmatis in spine implant infection
A 62-year-old female with history of osteoarthritis, hyperlipidemia, depression, and anxiety initially presented to clinic with approximately two years of frequent falls and progressively worsening issues with balance and coordination. She denied any neck or back pain, but did note weakness, numbness, and tingling to upper extremities, with symptoms worse on the right side. She had begun to require the use of a walker for ambulation, and saw no relief with multiple epidural injections or aqua therapy. Physical exam revealed decreased range of motion of her cervical spine, and decreased strength to bilateral triceps, wrist flexors, and intrinsics. CT and MRI of the cervical spine demonstrated myelopathy and destructive changes consistent with advanced osteoarthritis at C1-C2, and the patient subsequently underwent a C1 to C3 laminectomy and occiput to C5 posterior fusion.
At one-month follow-up, the patient reported intermittent posterior neck pain and frontal headaches, but overall her symptoms had improved. However, at 42 days status-post her initial surgery, she presented to clinic with a week of shooting pain up her posterior head, and radiographic imaging revealed two occipital screws displaced and migrated from the plate. As the patient showed no constitutional signs or symptoms of infection, she was indicated for a revision surgery with placement of new instrumentation. However, during the procedure, there was noted to be a large amount of inflammatory tissue surrounding the disengaged screws, and three samples of this tissue were sent for analysis with culture.
Culture of tissue grew a nontuberculous mycobacteria (NTM) in five days, but a specific species was not identified using culture until five weeks following sample collection. In contrast, next-generation sequencing (NGS) analysis performed on samples obtained during a subsequent irrigation and debridement confirmed the presence of Mycobacterium smegmatis within one week. Although traditional culture identified a rapidly growing NTM in this case of spine implant infection (SII), the species was not identified in a timely manner. Accurate and rapid species-level pathogen identification is imperative in the targeted treatment of spinal infections including those caused by NTM. The use of molecular testing such as NGS is helpful in identifying infecting organisms and guiding antimicrobial treatment.
Chronic sinusitis requiring dual antibiotic therapy
A 57-year-old male with longstanding chronic sinusitis and history of prior endoscopic sinus surgery presented with an exacerbation of his chronic sinusitis. A sinus aspirate was obtained and MicroGenDX next-generation DNA sequencing performed. This revealed medium quantities of bacterial DNA. Three facultative anaerobic organisms were identified: Enterobacter cloacae, Corynebacterium segmentosum, and Staphylococcus epidermidis. The Enterobacter species was found to represent 61% of the bacterial DNA, and the Corynebacterium represented 34%, while S. epi represented only 2% of the DNA – confirming it to be a contaminant. To adequately treat the infection, and given the lack of oral antibiotic coverage with a single agent for both species, dual antibiotic therapy with both doxycycline and ciprofloxacin was used. The patient had complete resolution of infection with one 14-day course of therapy.
This case highlights two important advantages of next-generation DNA sequencing for microbial identification in patients with chronic sinusitis. First, although three bacterial species were identified, the quantitative nature of the results allowed for determination of the relative contribution of each species to the infection, including determining that S. epi was a contaminant. Second, by identifying more than one organism, the need for combined antimicrobial therapy was established, and a treatment with both ciprofloxacin and doxycycline resulted in rapid resolution of infection. The selection of ciprofloxacin and doxycycline would certainly not have occurred as empiric therapy.
Patient with urinary tract infection
An 82-year-old woman with a history of recurrent UTI, and who had been treated for E. coli, presented to the clinic with an acute episode.
A mid-stream urine sample was obtained and sent for standard urinalysis, C&S, PCR by Volente Laboratories and MD Labs, and NGS by MicroGenDX.
There was a high concordance between C&S and PCR-based techniques to detect a high load of E. coli (>105 per ml). All testing identified E. coli, but NGS was the only lab that detected the dominant pathogen as Bacteroides fragilis (50%). Based on the anaerobic B. frag and quinolone-resistant genes detected by NGS, the patient was switched to a targeted antimicrobial therapy using clindamycin for a total of 14 days. By the fifth day, the patient’s symptoms subsided, and upon follow-up on day 14, she had no symptoms. View study
NGS finds fungus that culture missed to resolve life-threatening wound
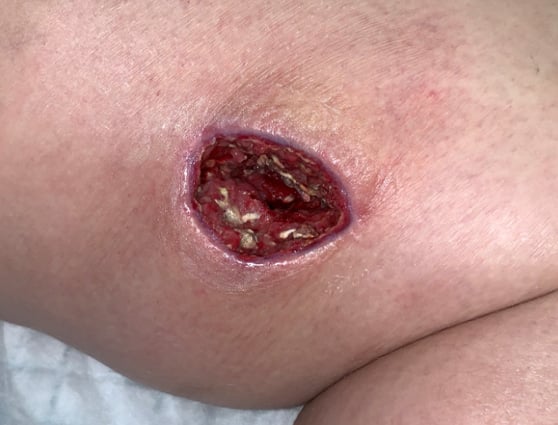
A young woman with type I diabetes, who had lost her kidney transplant a year previously, was hospitalized twice over six months with sepsis, the source of which remained a mystery. A large transplant surgery abdominal wound had been open for months, as had a heel pressure sore complicated by peripheral arterial disease, but both improved with better nutrition and regular wound care.
The patient then presented with sudden left leg pain, swelling, and the rapidly-ascending erythema characteristic of necrotizing infection. Fasciotomies of her lower leg were performed, and her left medial thigh was opened to drain what appeared to be an abscess. The muscle was perfectly pink and only clear, odorless fluid was found. This fluid then showed no growth on culture. Although wounds improved initially with NPWT, the patient didn’t look or feel better, and within a week the leg wound had grey/black tissue that appeared to be necrotic.
The same fluid was sent to MicroGenDX for DNA analysis (see full report), which detected Rhizopus — one of several fungal species that cause mucormycosis — a serious but rare fungal infection with a mortality rate of 50%. Early detection and treatment are critical for the best outcome, and the transplant team was notified immediately. Read study
