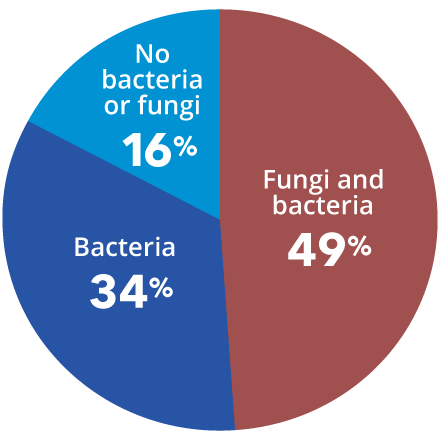
Consensus Guidelines Recommend DNA Diagnostic Testing During Phase I of Chronic Wound Management
DNA diagnostics recommended in first 1-4 days
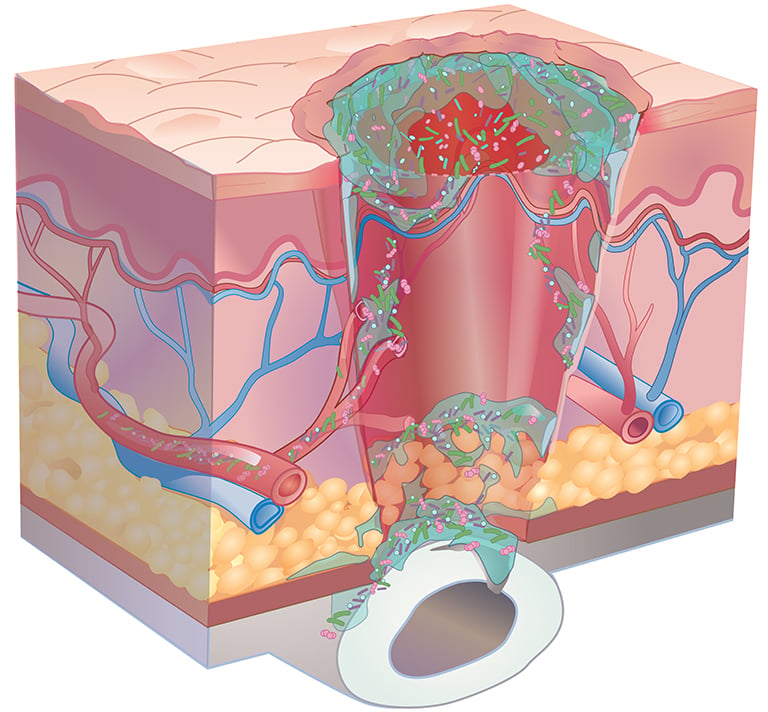
Consensus guidelines from a Global Wound Biofilm Expert Panel include the use of DNA diagnostics in the first 1-4 days of chronic wound therapy as part of early intervention. When all other healing measures are taken into account, there is strong evidence linking bacterial proliferation to chronic non-healing wounds. A sustained presence of proinflammatory agents degrades the extracellular matrix, inhibits cell migration, and prevents wound healing.
State-of-the-art qPCR + next-gen sequencing (NGS) increases precise species identification more than 20-fold when compared to culture. qPCR+NGS also provides an estimated percentage of the organisms present in the specimen. This specificity of organism identification and estimated quantification empowers the clinician to treat a patient with absolute confidence.
Published research shows accelerated healing with MicroGenDX
| Standard of Care Group | Group 1 | Group 2 |
|---|---|---|
| Traditional Culture with Systemic Antibiotics | Molecular Diagnostics with Systemic Antibiotics | Molecular Diagnostics with Customized Topical Antibiotics |
| 48.5% patients healed | 62.4% patients healed | 90.4% patients healed |
A more comprehensive microbial view affords the greatest potential of diminishing polymicrobial communities within this window. Based on a 1300+ patient study, the use of molecular-guided therapeutic choices resulted in substantial clinical improvements to both healing rates and healing days as compared to culture-guided choices.
MicroGenDX qPCR+NGS is superior to culture and PCR-only for diagnosis and treatment of chronic or stalled wounds
| Standard Culture | PCR Only | MicroGenDX qPCR+NGS |
|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
MicroGenDX qPCR+NGS also has superior diagnostic capabilities for nail infections
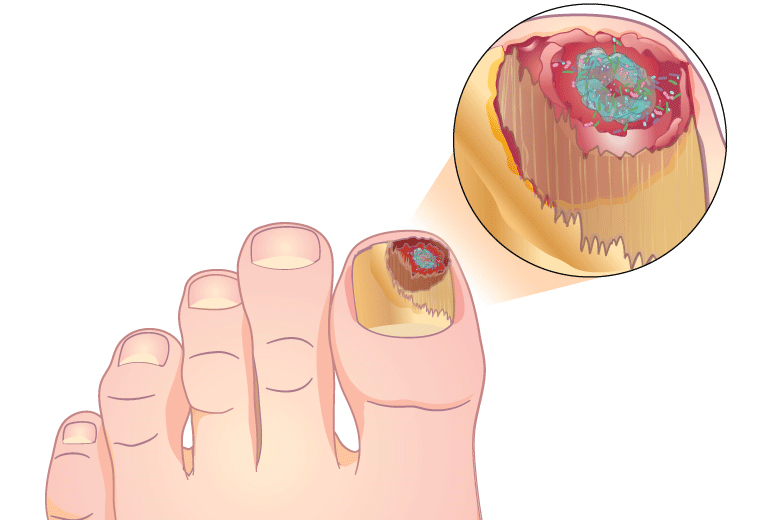
Combining qPCR and NGS offers greater speed and sensitivity, and more reliable speciation, than PAS/GMS or culture.
“As clinicians we can all generally agree that in order to provide the best treatment, we need to understand the organism that we’re dealing with. With MicroGenDX DNA sequencing, we can do this for nail pathogens and create a targeted DNA therapy that is specific to the nail.” — Beth Pearce, DPM
| NGS | qPCR | PAS/GMS | Culture |
|---|---|---|---|
|
|
|
|
References
- van Asten S A V, La Fontaine J, et al. The microbiome of diabetic foot osteomyelitis, European Journal of Clinical Microbiology & Infectious Diseases. 2016; 35):293-298. doi: 10.1007/s10096-015-2544-1
- Joyce A, Gupta A, et al. Fungal Diversity and Onychomycosis – An Analysis of 8,816 Toenail, Journal of the American Podiatric Medical Association. 2019 Jan;109(1):57-63. doi: 10.7547/17-070
- Chadwick P. Fungal infection of the diabetic foot: The often ignored complication. The Diabetic Foot Journal 16: 102–7, 2013
- Dowd S E et al Molecular Diagnostics and personalized medicine in wound care: assessment of outcomes. Journal of Wound Care. 2011; 20: 5, 232-239
- 2009-2020 College of American Pathologists proficiency data

 Chronic wounds
Chronic wounds Nail Infections
Nail Infections